The analysis of the large amount of data generated by sequencing technologies as RNAseq and ChIPseq is computational demanding and it is usually associated with the need of reproducibility of the analysis performed.
Reproducibility demands for a “freezing” of the software used in the analysis, which is not easy to achieve when the analysis is outsourced to third parties, e.g. clouds based applications.
To address this aspect, we provide a cheap, efficient and reproducible data analysis ecosystem based on NUC6I7KYK mini-PC (Fig. A – panel 4: SeqBox hardware), and on Docker platform.
This solution, namely SeqBox, supports, via a friendly GUI, the analysis of RNAseq and ChIPseq data (Fig. A - Panels 1-3: SeqBox software), thus allowing a wide access to fast and reproducible analysis to scientists with/without coding experience.
An example of SeqBox assembly

Fig. B
(1) SeqBox,
(2) SeqBox power supply,
(3) External data HD NTFS formatted, not provided,
(4) PC/MAC connected to SeqBox via Ethernet cable, not provided,
(5) Ethernet cable, not provided.
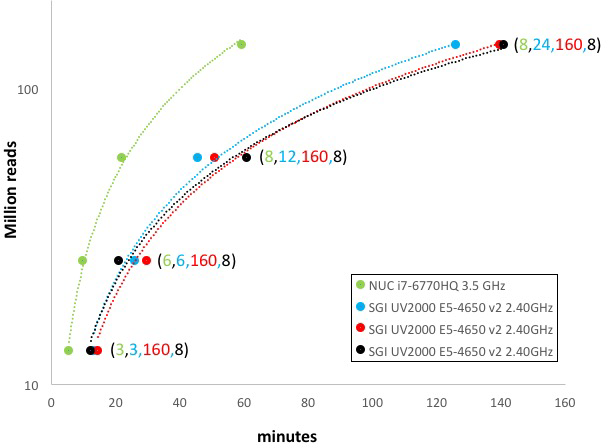
Performance comparison between SeqBox and SGI UV2000 server (Fig. C)
Numbers in parenthesis indicate the number of cores/threads assigned for parallelization, e.g. 8 refers to the number of cores associated to the NUC miniPC.
Further information on SeqBox
- About the SeqBox ecosystem
- RNAseq workflow (textual description)
- miRNAseq workflow (textual description)
- ChIPseq workflow (textual description)